Introduction
Because of simplicity and proven success in industrial process design, the Taguchi methods (Taguchi 1986, Fowlkes and Creveling 1995) have recently offered a cost-effective strategy for the study of interactions between reaction variables in DNA amplification (Cobb and Clarkson 1994, Caetano-Anollés 1998). Taguchi methods provide accurate prediction of component levels for optimum performance, and as such, they have been widely used in the automotive and electronics industries with success and are now being extended to the food and pharmaceutical sectors (Charteris 1992). These methods use quadratic loss functions (signal-to-noise ratios) that penalize deviations from prediction values and orthogonal array designs capable of examining many factors in only few experimental trials. Using Taguchi methods, a set of 8 variables (k) can be arranged in an orthogonal array that defines only 18 amplification reactions (2k + 1, or next multiple of 3; where k=8). Instead, a full factorial design of 8 variables studied at 3 levels (e.g., concentrations) demands an experiment with 6561 (3 exp8) separate reactions. In this section we will introduce the Taguchi method as a general optimization tool for many methods in molecular biology. We will describe the optimization of a complex nucleic acid scanning technique using this strategy for robust industrial design and as recently described (Caetano-Anollés 1998). However, other amplification or hybridization-based methodologies that depend on a number of variable factors could equally benefit from the application of this powerful optimization approach. Return
Methods
1. Define your experimental system. For example, Caetano-Anollés (1998) based the DAF optimization exercise on the amplification of soybean (Glycine max L. cvs. Bragg and Minsoy) and flowering dogwood (Cornus florida L. cvs. Cherokee Brave and Appalachian Spring) with a group of four octamer primers (GTAACGCC, GACGTAGG, GAAACGCC and GTATCGCC).
2. Define the experimental variables (parameters). For example, variables can include enzyme, primer, template and magnesium chloride concentration, annealing, denaturation and extension temperature and time, and number of cycles.
3. Scan electrophoretic gels (e.g., silver stained DAF gels below) and analyze acquired images using the program NIH Image (v. 1.61) or any other image analysis software.
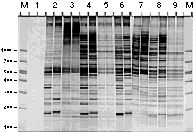
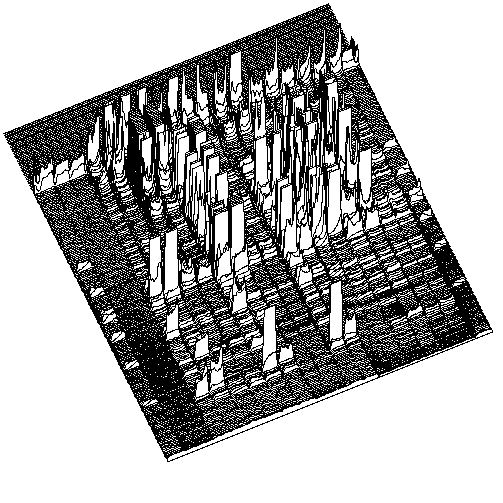
4. Obtain quantitative estimates of amplification efficiency and multiplex ratio. For example, amplification yield (I) values can be obtained by integration of relative peak areas from density profiles generated from the entire surface of individual fingerprints. Product number (N) can be defined in many ways; eg. N = m + 1, where m was the scorable number of amplification products of less than 1000 bp in length.
5. Evaluate the interaction of parameters using for example L9 (3 exp4) and L18 (3 exp8) orthogonal arrays capable of examining 4 and 8 factors (variables) at 3 levels each, respectively (Taguchi 1987).
6. Duplicate experiments in a crossed array layout to control for noise factors (Fowlkes and Creveling 1995), each set of tubes assembled independently and amplified separately. The layout can control noise from block heating inhomogeneities, errors in reaction assemblage (e.g., pipetting, reagent concentration differentials), and silver staining variability.
7. Use individual values (y) of either I or N corresponding to each of 9 or 18 amplification runs to calculate 'larger-the-best' (LTB) signal-to-noise ratios (S), according to the formula:
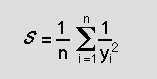
where n is the total number of individual data points scored. S values can also be calculated using a modified Taguchi pooled-level method (Cobb and Clarkson 1994) which estimates S for each level as the average of values from individual runs and defines n as the number of levels.
8. Calculate factor effects by analysis-of-means (AOM), averaging S for each control factor level.
9. Calculate optimum levels for each factor as those that maximized S, and infer their value by polynomial regression (p = mx2 + mx + c).
9. Calculate S values for the mean of the overall experiment (Sexp) and for the optimum configuration (Sopt). Used these values in a verification test (Fowlkes and Creveling 1995). Use analysis of variance (ANOVA) to decompose the variance for each factor of parameter design and identify the strongest contributing control factors. Return
An exercise on DAF optimization
During the past few years, arbitrarily amplified DNA (AAD) techniques such as RAPD, DAF and AFLP have been successfully used in numerous and diverse applications that include mapping and tagging of traits and the study of biological diversity. The use of AAD markers depends on amplification components and thermal cycling parameters, and its reproducibility and correct usage is contingent on adequate optimization. This demands an investigation of the interaction between many variables and involves unusually large experiments. Taguchi methods have been used efficiently to optimize AAD analysis and study interaction between amplification and thermal cycling parameters (Caetano-Anollés 1998). Optimized parameters defined robust and transportable protocols capable of producing reproducible results both within and between laboratories.
The Taguchi strategy for robust design was first used to optimize reaction components in DAF analysis (Caetano-Anollés 1998). A number of progressive trials, based on L9 (3 exp4) orthogonal arrays and 9-reaction tube experiments, identified variables that had major effects on amplification yield and product number. For each component, optimum conditions were those that maximized the estimated type B quadratic loss function (S) values in Taguchi LTB analysis. Initial experiments defined "reproducibility windows" for the most important factors, i.e., the range of component levels over which no major variation occurred in number, distribution and intensity of amplified products. These factors were again studied using the standard Taguchi approach (Taguchi 1996) or a pooled-level modification (Cobb and Clarkson 1994), defining optimum reaction component levels (7-8.5 µM primer, 2.9-3.9 mM magnesium, and 0.5-0.7 units enzyme /µl). Inferred optima coincided or were slightly lower than those recently identified in DAF analysis of pathogenic fungi using high annealing temperatures (Bentley and Bassam 1996). Overall analysis of quality metrics for robust design showed that high primer concentration and annealing temperature established the complex nature of DAF patterns, enzyme concentration accounted for most of the variation observed in the amplification response (p<0.01), enzyme and magnesium concentration fine tuned the number of amplified products; and marked deviations from optimum levels were tolerated due to wide reproducibility windows.
Thermal cycling parameters were also studied (Caetano-Anollés 1998). Using optimized reaction components, eight thermal cycling factors were analyzed in a large L18 (3 exp8) orthogonal array-based Taguchi optimization study of amplification yield and product number. Experiments were independently replicated to control for experimental noise and used 4 octamers to account for the effect of primer sequence. Mean values (ÿ) and S values were calculated for each treatment and primer as well as for the overall experiment. A factors control table was constructed using AOM average S values corresponding to those reaction tubes that had a same control factor level. This defined three S ratios for each control factor, level, and primer with which to determine optimal factor levels by polynomial regression. Control factor effect plots ( see Figure) showed that annealing temperature and time were the most important control factors.
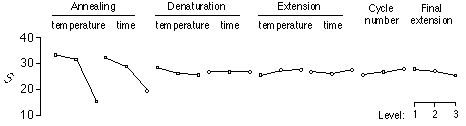
In contrast, there was little variation observed in the other thermal cycling parameters, which acted as weak control factors. These results were further clarified in ANOVA studies that decomposed the contribution of each individual control factor to the total experimental response (see ANOVA Table). Analysis of the overall experiment showed that annealing temperature and time were highly significant (F>4) and accounted for 66% and 29% of the overall response, respectively. In these experiments, variability due to primer was up to 100 times greater than due to experimental noise. However, such variability did not obscure the strong effects and interaction of annealing temperature and time.
|
Factor |
SS |
df |
MS |
F |
rho |
|
Annealing temperature |
1157.9 |
2 |
578.9 |
123.2** |
65.6 |
|
Annealing time |
516.4 |
2 |
258.2 |
54.9** |
29.0 |
|
Denaturation temperature |
26.8 |
2 |
13.4 |
2.8 |
1.4 |
|
Denaturation time |
0.1 |
2 |
0.06 |
_ |
0 |
|
Extension temperature |
15.2 |
2 |
7.6 |
_ |
0.8 |
|
Extension time |
6.8 |
2 |
4.4 |
_ |
0.3 |
|
Cycle number |
14.6 |
2 |
7.3 |
_ |
0.7 |
|
Final extension |
10.1 |
2 |
5.0 |
_ |
0.5 |
|
Pooled error |
46.8 |
(10) |
4.7 |
||
|
Error |
2026.5 |
54 |
37.5 |
The sum of squares (SS) quantifies the variation induced by a factor around the overall experimental mean response, and the factor mean square (MS) estimates is variance. The error variance was calculated from the SS due to experimental error and was determined by replication.
F values were defined by using error variance calculated by pooling SS values from control factors that have a small contribution to the overall S ratio. **, significant at the 99% confidence level.
df, degrees of freedom. rho, contribution ratio (%).
Regression analysis defined optimum levels for each variable analyzed. These levels were comparable to those obtained using the modified Taguchi approach or when analyzing product number. Congruence indicates that there is no inherent bias introduced by the way how S ratios were calculated from product yield or number. However, in some cases there were marked differences in value (cf. annealing and denaturation time). Annealing temperature was optimum within the 45-48°C range depending on the primer used or whether optimization was centered on number or yield of amplified products. Similarly, optimum annealing times were within 60-130 s. Optimum factor set points maximized S over any S calculated for individual treatment combinations. Using these maximized S values and the overall experimental average (Sexp=26.8), optimum S ratios (Sopt=44.0) were calculated from a predictive equation (Fowlkes and Creveling 1995). Predicted Sopt ratios were then compared to experimental ratios (Stest=44.5) obtained in a set of verification experiments whereby DNA was amplified using optimized conditions. This comparison confirmed the validity of inferred optima since observed Stest ratios were in good agreement with those inferred by Taguchi analysis. This analysis established that optimum conditions were predictable, verifiable and reproducible, and that the experimental design was intrinsically robust.
The optimized amplification protocol was finally tested in its transportability and inter-laboatory use. Three thermocycler units that differed widely in response time to temperature equilibration were compared in these studies, and the reproducibility of the Taguchi-optimized DAF protocol was confirmed by replicate amplification of soybean DNA in 3 separate experiments based on the different thermocyclers. The protocol was tested in the analysis of a number of plant templates, including dogwood, soybean, chickpea (Cicer arietinum L.) and bermudagrass (Cynodon sp.), and in the amplification of the pathogenic fungus Discula destructiva. It has shown to be robust and highly reproducible, probably due to the existence of wide reproducibility windows that tolerate variations introduced by factors such as template, pipetting error, concentration differences in reagent stocks, and temperature gradients during thermal cycling. Return
WinRobust program for robust experimental design
WinRobust is a program for IBM PC compatible computers (running Windows) capable of assisting both the engineer and the scientist in the planning and analysis of Quality Engineering experimentation. The program assumes the user has reasonable knowledge in the practice of Quality Engineering, Robust Design and Taguchi Methods. WinRobust automates the laborious tasks of assigning factors to orthogonal arrays, calculating signal-to-noise ratios, and producing factor effect plots. The program can be purchased from Abacus Digital Products (6 Lookout View Rd., Fairport, NY 14450) or from ColorPoint Graphics (274 N Goodman St., Rochester, NY 14607). Return

